Glossary
| Term | Definition |
|---|---|
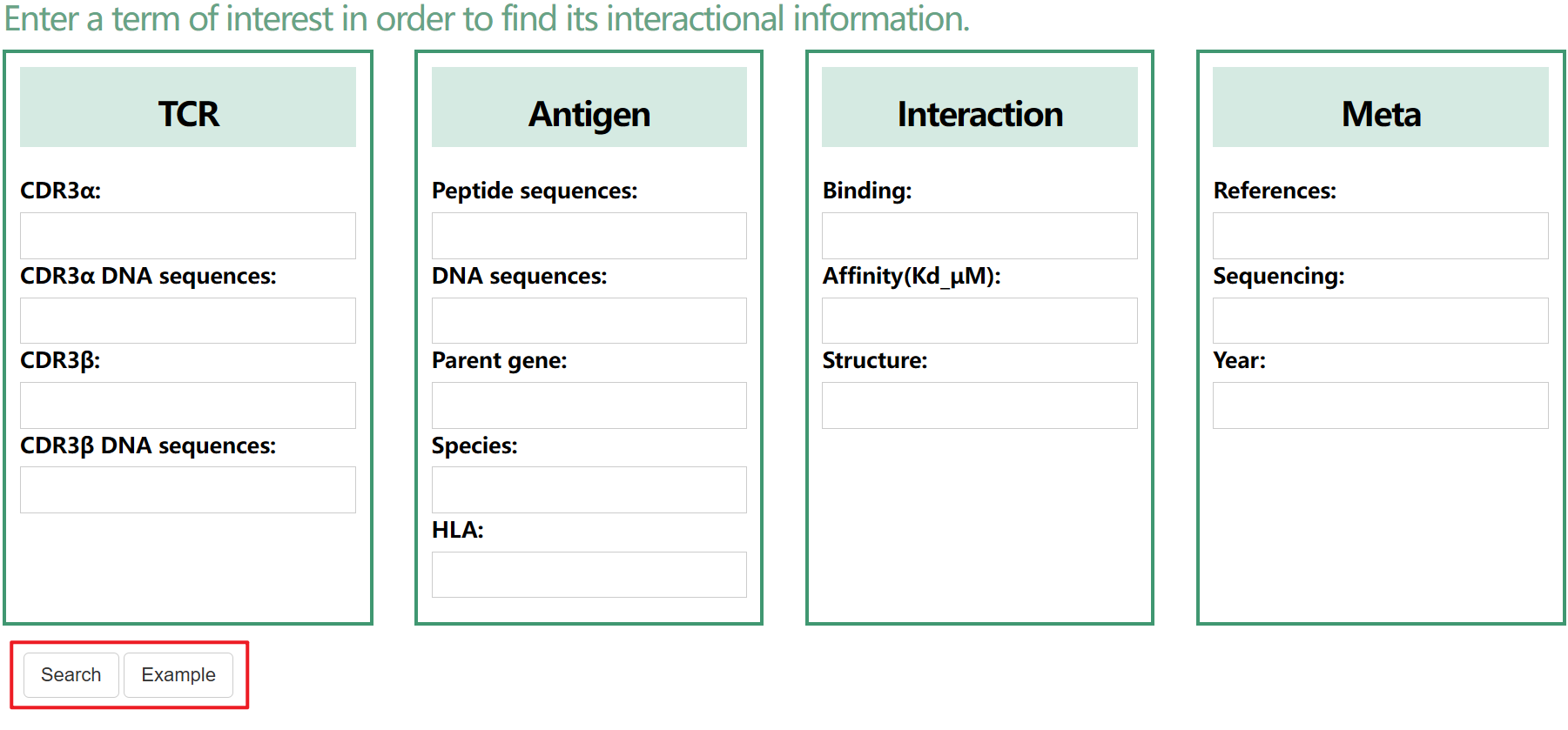
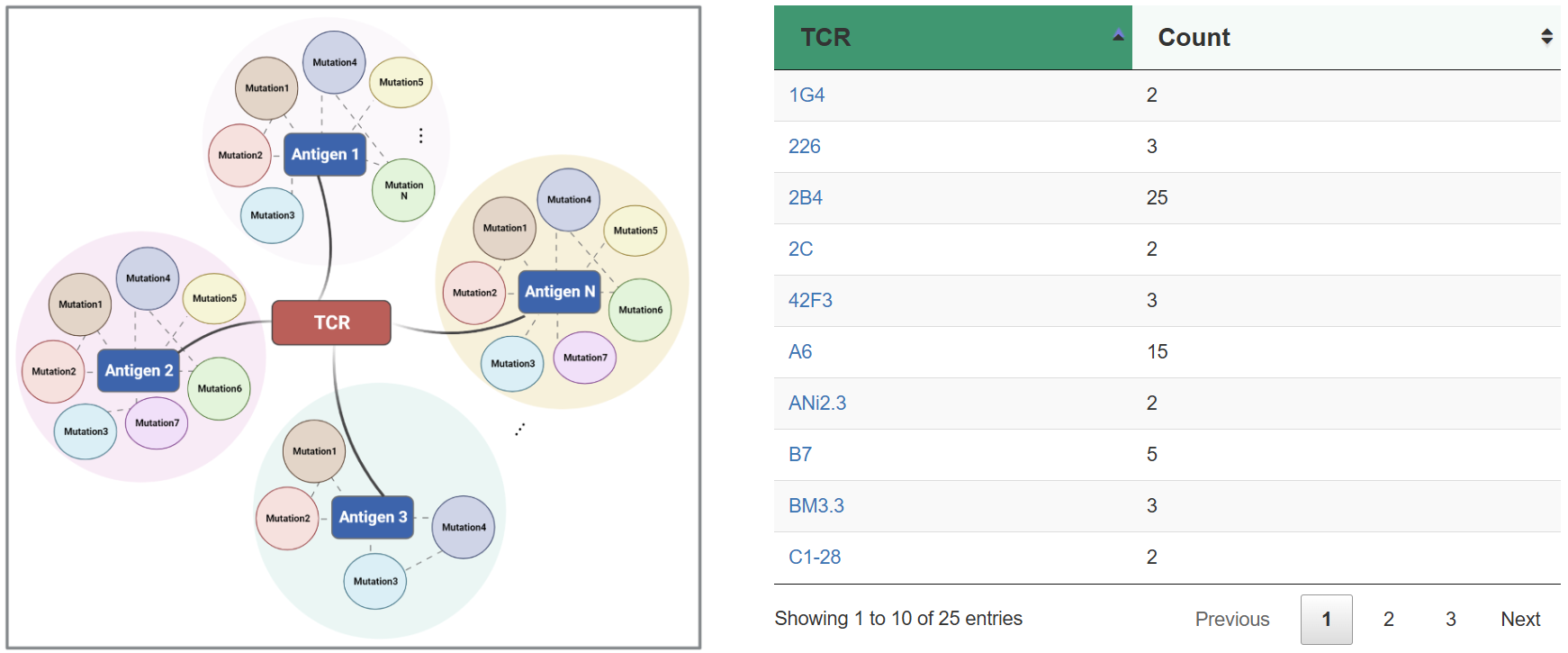
| TCR | T cell receptor. |
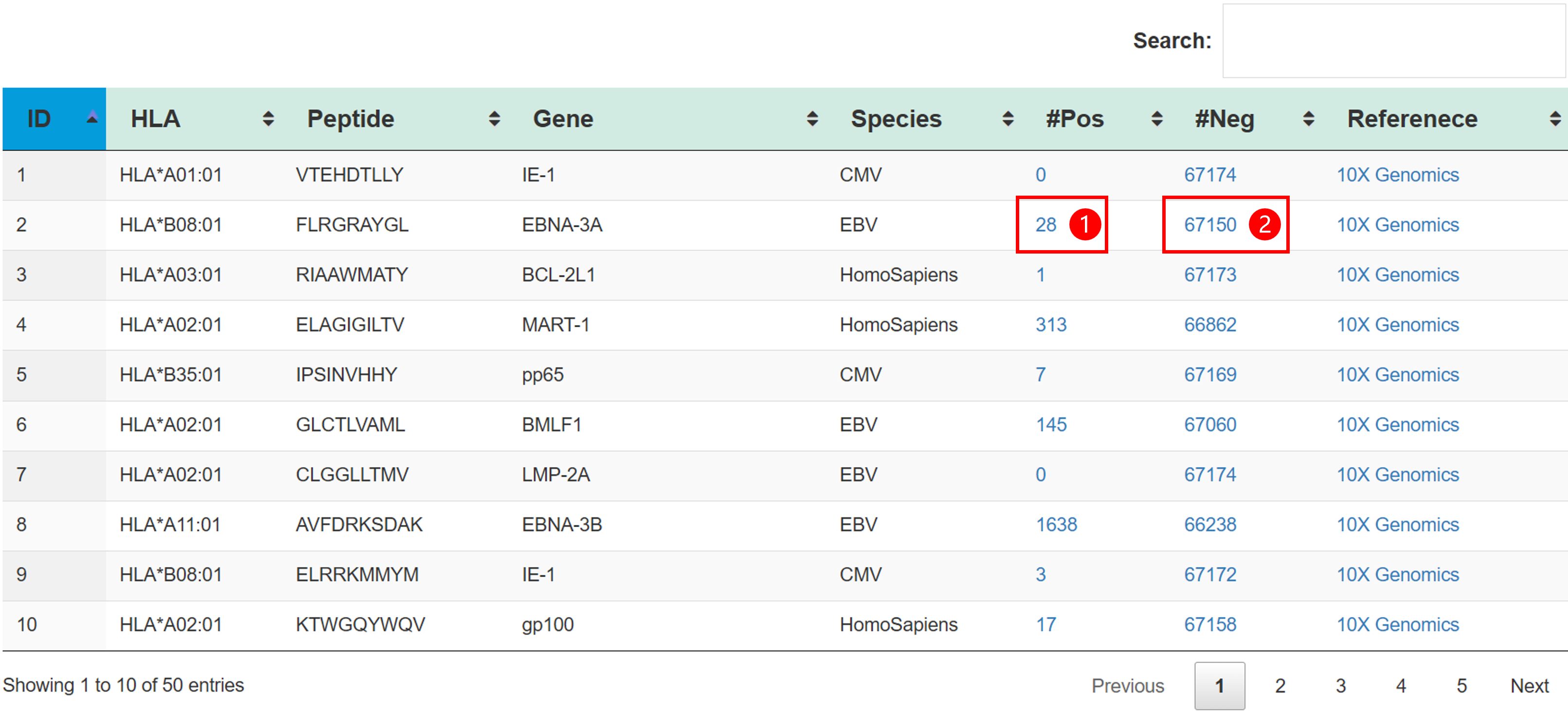
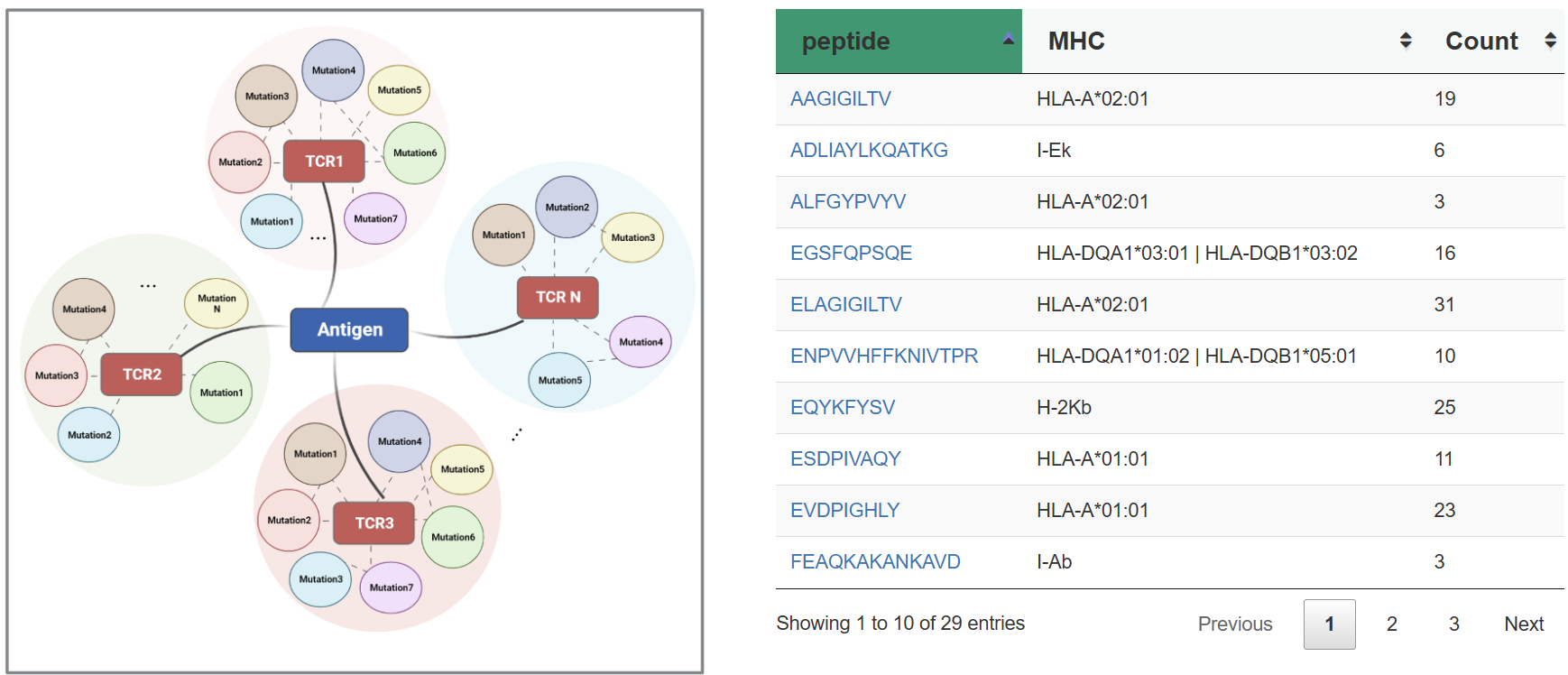
| Antigen | Peptide-major histocompatibility complex multimer which can be recognized by TCR. |
| MHC | The major histocompatibility complex. |
| HLA | The human leukocyte antigen. |
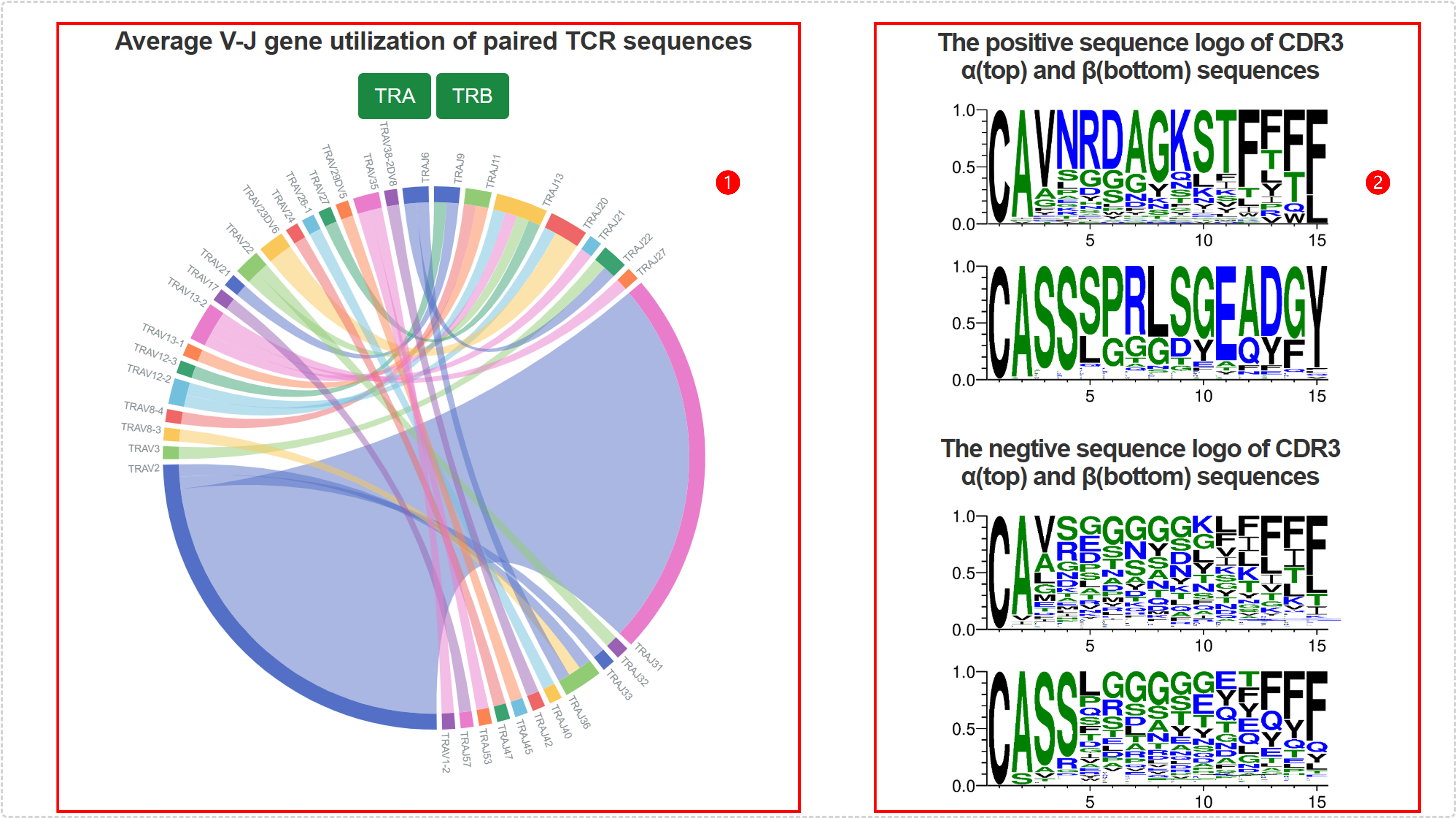
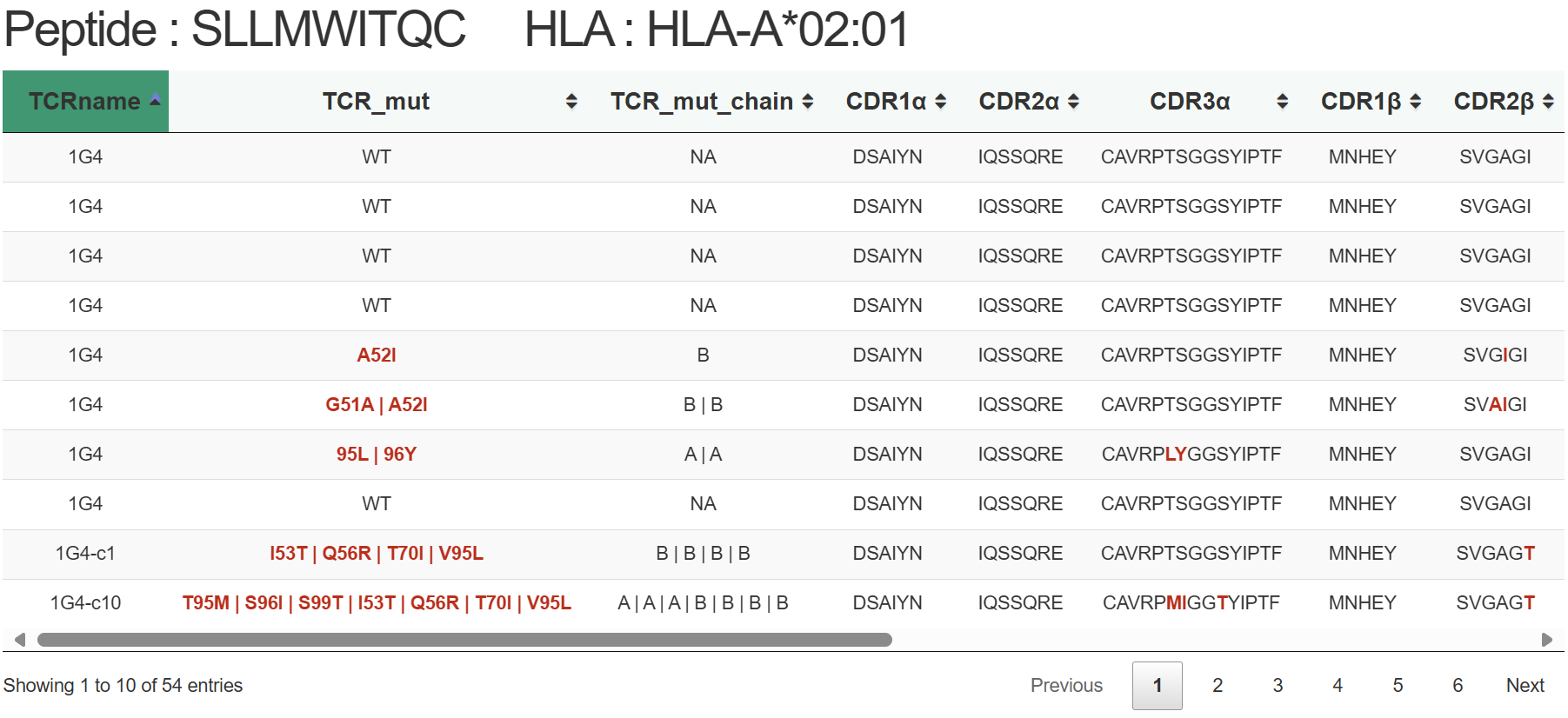
| CDR3 α/β | The third complementarity determining region of the T-cell receptor(TCR) α and β chains. |
| TRAV/TRAJ | V/J gene of TCR α chain. |
| TRBV/TRBJ | V/J gene of TCR β chain. |
| Peptide_species | Species of peptide. |
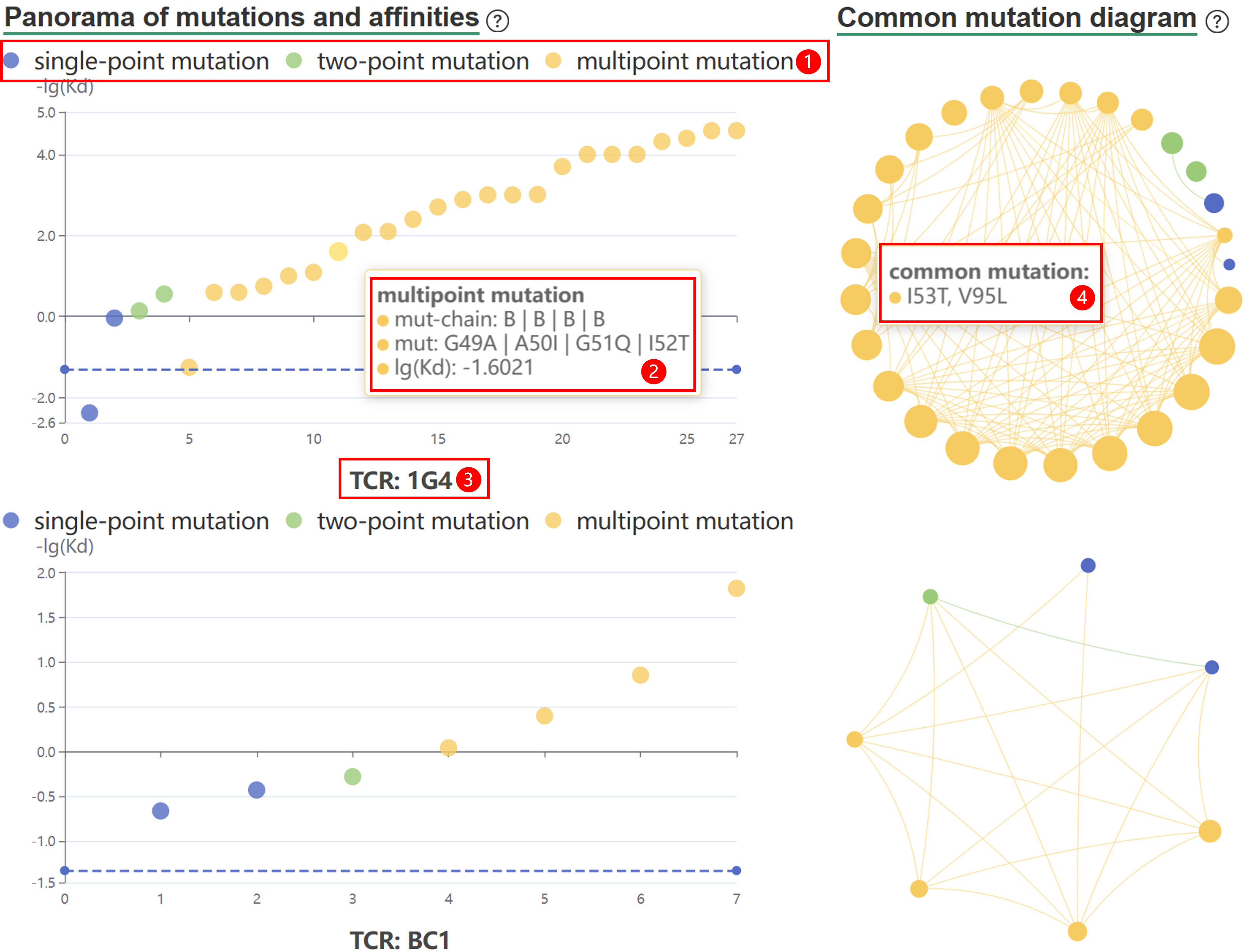
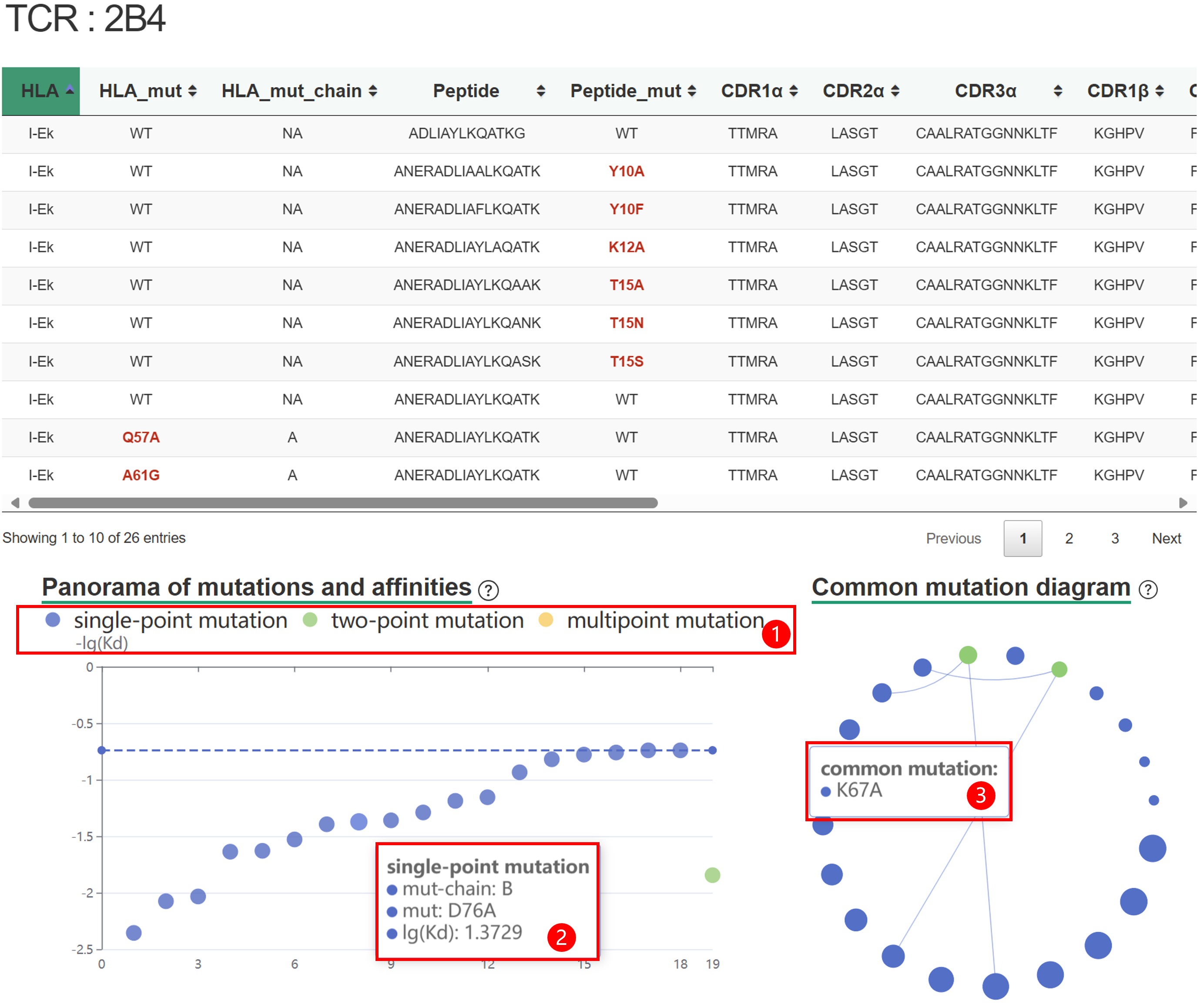
| HLA_mut/Pep_mut/TCR_mut | The mutant site of HLA/peptide/TCR. |
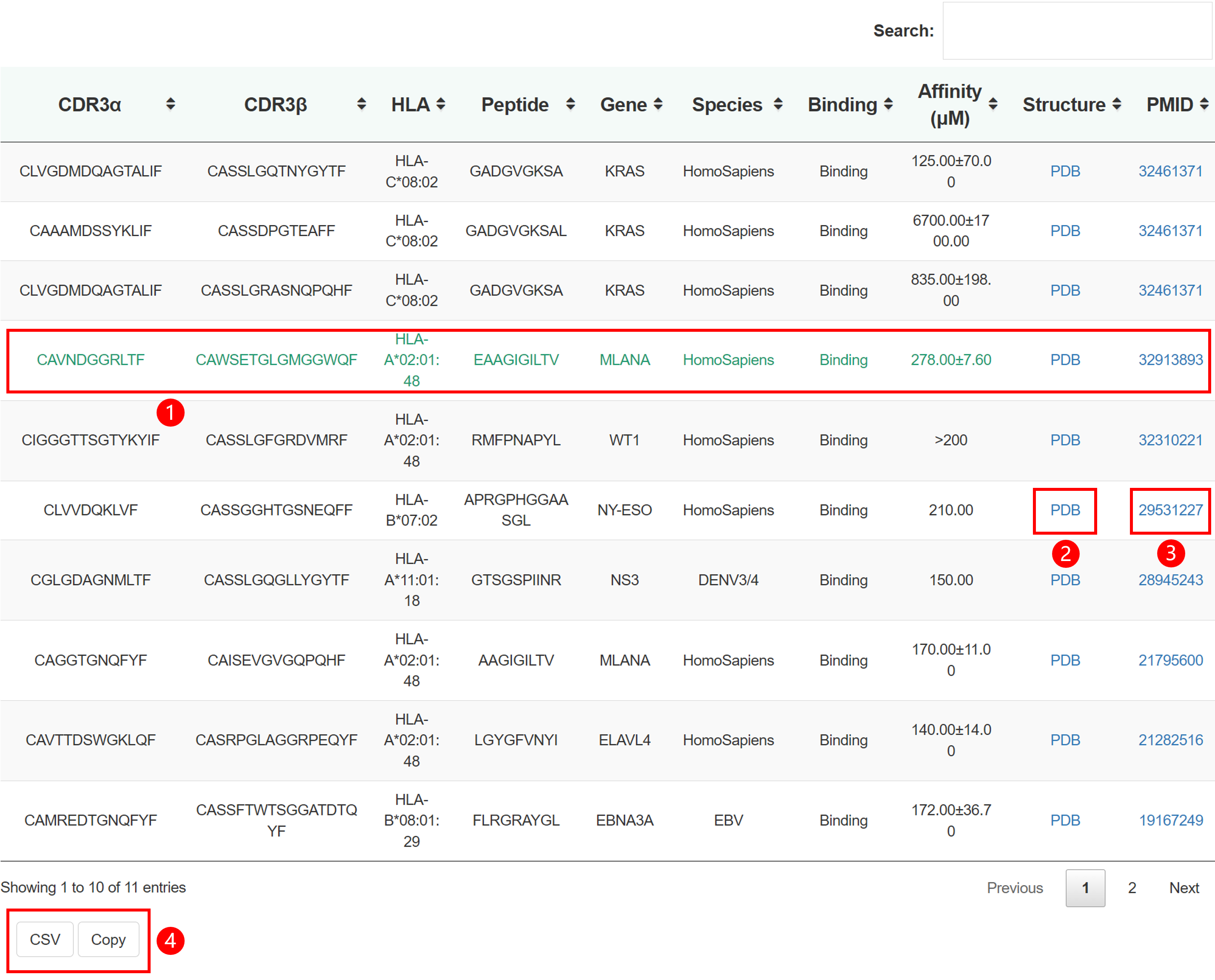
| Binding | Descript the binding capacity between TCR and pMHC. TRUE (Binding); FALSE(Non-Binding). |
| Affinity | The binding affinity between TCR and pMHC. |
| Structure | The structure of TCR-pMHC complexes from PDB. |
| Interactive pairs | The pairs of TCR and antigen that can interact with each other. |
| Non-interactive pairs | The pairs of TCR and antigen that can not interact with each other. |
Usage of TRAIT
TRAIT provides a landscape of experimentally verified interactive and non-interactive TCR-antigen pairs, together with multi-dimensional perspectives of interaction features integrating sequences, structures, and affinities. TRAIT is dedicated to providing users with relevant data from “discovery, optimization, and clinical evaluations” TCRs, facilitating the development of T cell-based immunotherapies. Some images on the webpage were created with BioRender.com. The main sections of this website are Home, Search, Omics, Mutations, Therapeutics, Download and Help.
About Us
Citation
Mengmeng Wei, Jingcheng Wu, Shengzuo Bai, Yuxuan Zhou, Yichang Chen, Xue Zhang, Wenyi Zhao, Ying Chi, Gang Pan, Feng Zhu, Shuqing Chen, Zhan Zhou, TRAIT: A Comprehensive Database for T-cell Receptor–Antigen Interactions, Genomics, Proteomics & Bioinformatics, 2025, qzaf033, https://doi.org/10.1093/gpbjnl/qzaf033
Contact
If you have any questions and helpful suggestions, please contact us.
E-mail: zhanzhou@zju.edu.cn
Address: College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, China.
Address: College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, China.
Updating
Latest Updates
1. TRAIT was last updated on March 16, 2025.
2. Affinity data has been updated on both the Search and Mutations pages.
2. Affinity data has been updated on both the Search and Mutations pages.