About
Citation
Please cite us if you find this resource useful:
[1]. Wu J, Zhao W, Zhou B, Su Z, Gu X, Zhou Z*, Chen S*. TSNAdb: a database for tumor-specific neoantigens from immunogenomics data analysis. Genomics Proteomics Bioinformatics. 2018, 16(4),276–282. DOI: 10.1016/j.gpb.2018.06.003 
[2]. Wu J, Chen W, Zhou Y, Chi Y, Hua X, Wu J, et al. TSNAdb v2.0: the Updated Version of Tumor-specific Neoantigen Database. Genomics Proteomics Bioinforma 2022:in press.
Data Source
Prediction Methods
The neoantigens are predicted by NetMHCpan v4.0,MHCflurry and DeepHLApan. NetMHCpan is the most frequently used tool for neoantigen prediction in clinical practice. MHCflurry obtains the prediction neoantigen efficiently and with high quality. DeepHLApan considers both HLA-peptide binding and immunogenicity of pMHC that the other two tools haven’t taken into consideration for high-confidence neoantigen prediction.
Databse usage
Basic information

The displayed data of 'Browse' page and 'Search' page are derived from the following process. Only the HLA-peptide pairs that meet all the criteria of three tools would be considered as potential neoantigens.
'Home' page
1. The related data would appear or disappear once click on it.
2. The detailed information of the tumor type would appear once click on it, it's same with the tumor type of 'Browse' page.
3. It's flexible to display one to 16 tumor types as users wanted .
'Browse' page
Mutation type
The information of 'Detailed neoantigen' would be discussed in the 'Search' section.
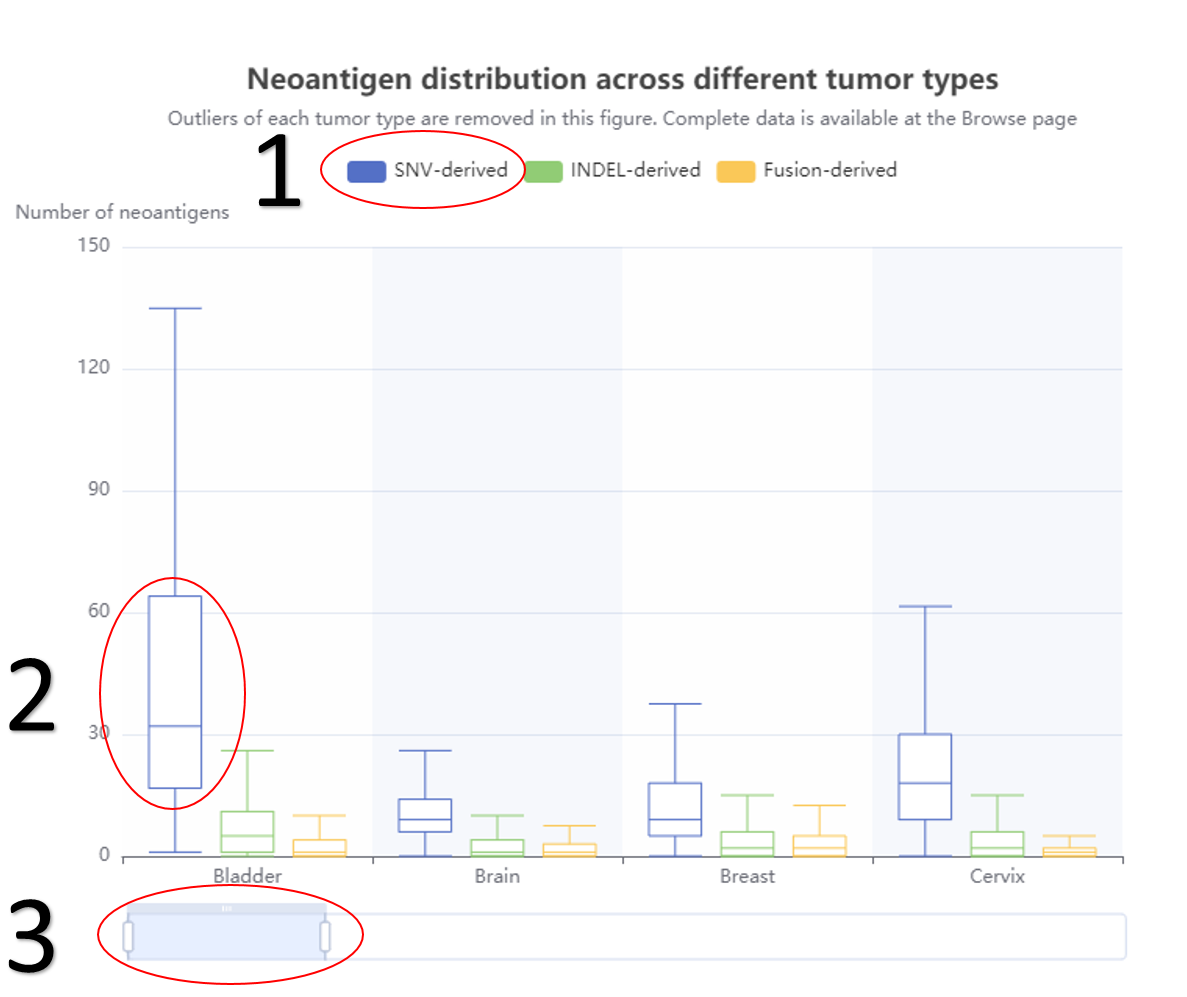
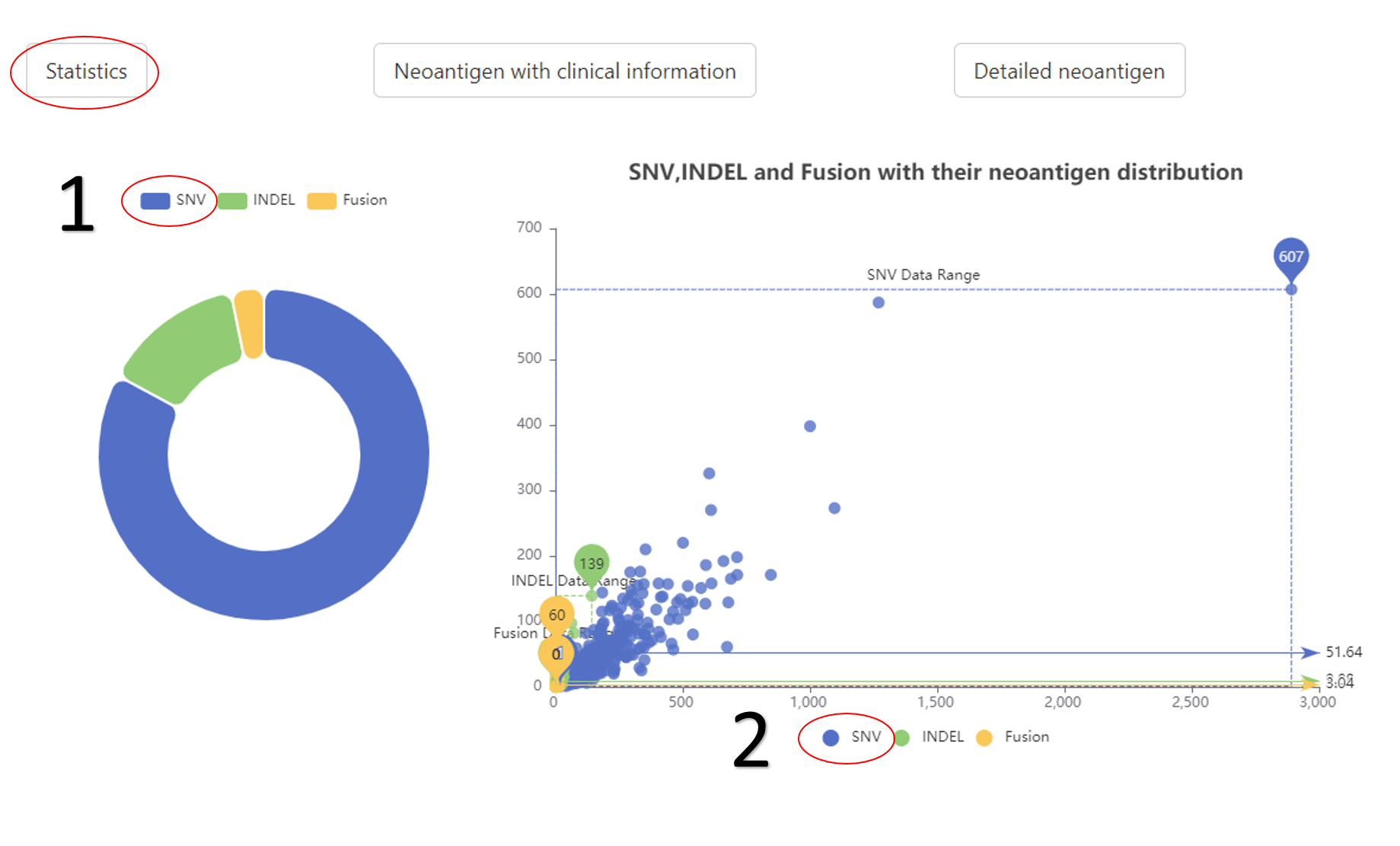
The statistical results of different mutation types.
1. The data distribution of neoantigen or mutation.
2. The related data would appear or disappear once click on it.

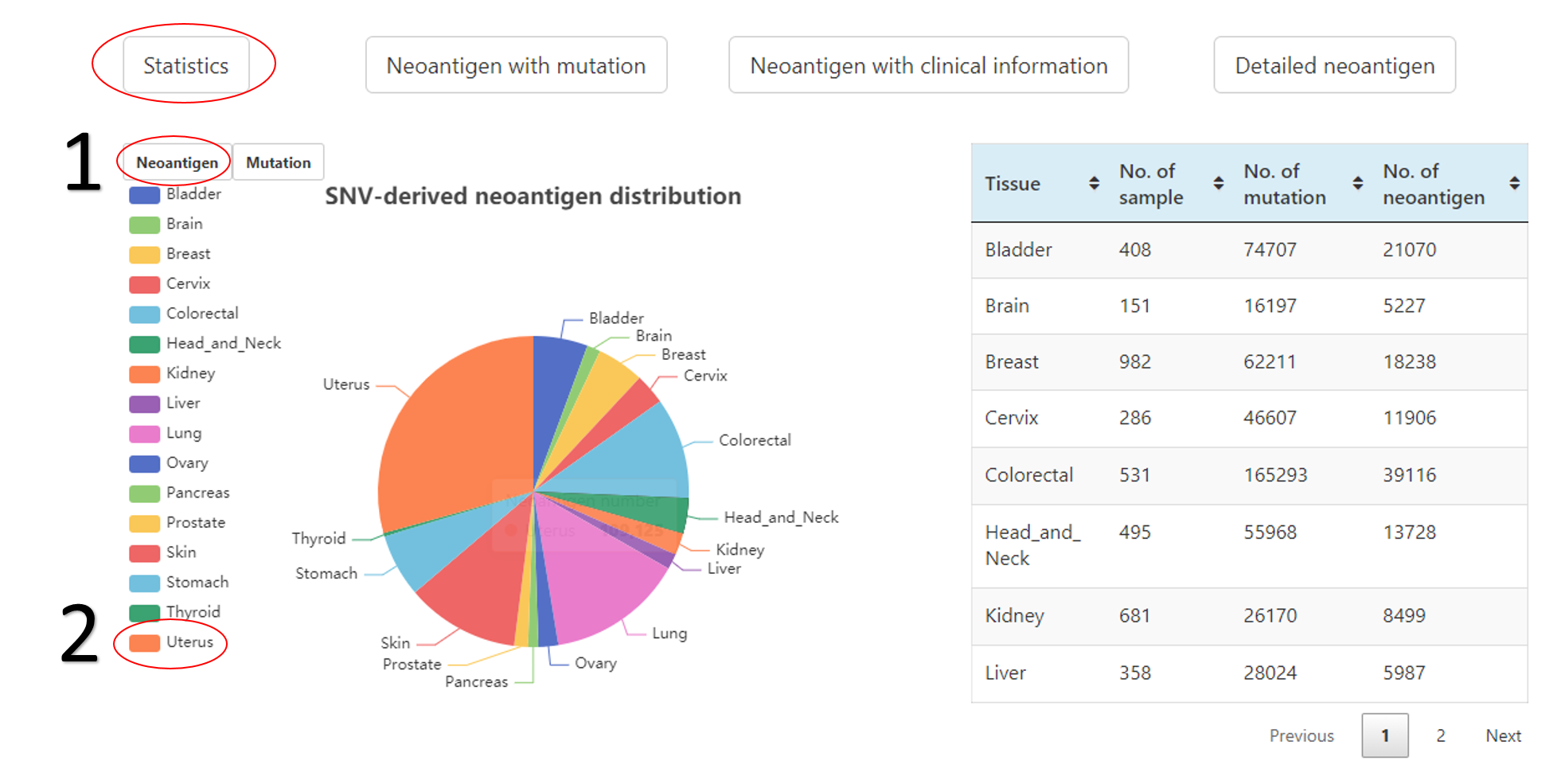
The relationship between neoantigens and mutations.
1. The related data would appear or disappear once click on it.
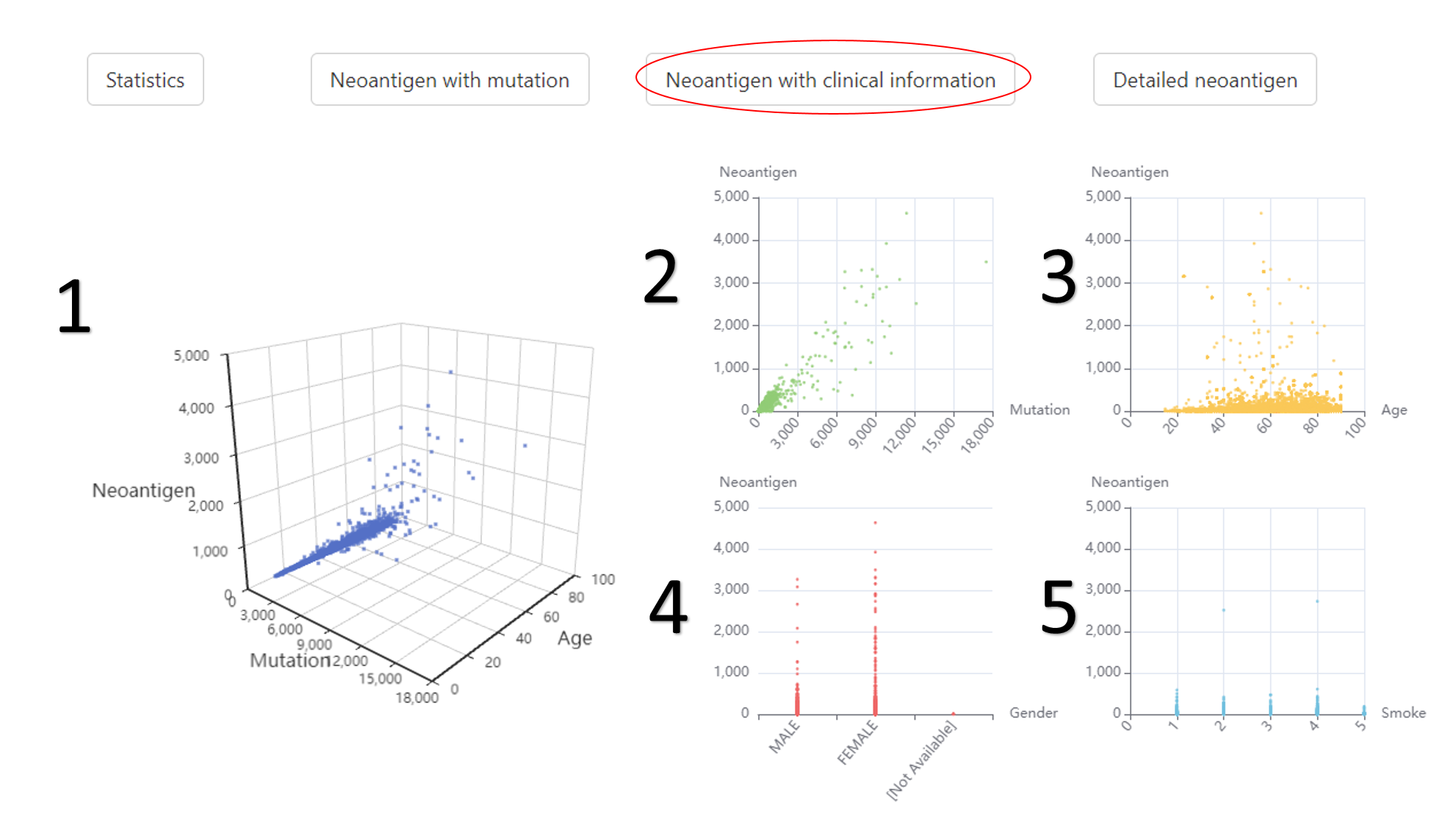
The relationship between neoantigens and clinical information.
1. The relationship among neoantigen, mutation and age.
2. The relationship between neoantigen and mutation.
3. The relationship between neoantigen and age.
4. The relationship between neoantigen and gender.
5. The relationship between neoantigen and smoke.
1: Lifelong Non-smoker (less than 100 cigarettes smoked in Lifetime)
2: Current smoker (includes daily smokers and non-daily smokers or occasional smokers)
3: Current reformed smoker for > 15 years (greater than 15 years)
4: Current reformed smoker for ≤15 years (less than or equal to 15 years)
5: Current reformed smoker, duration not specified
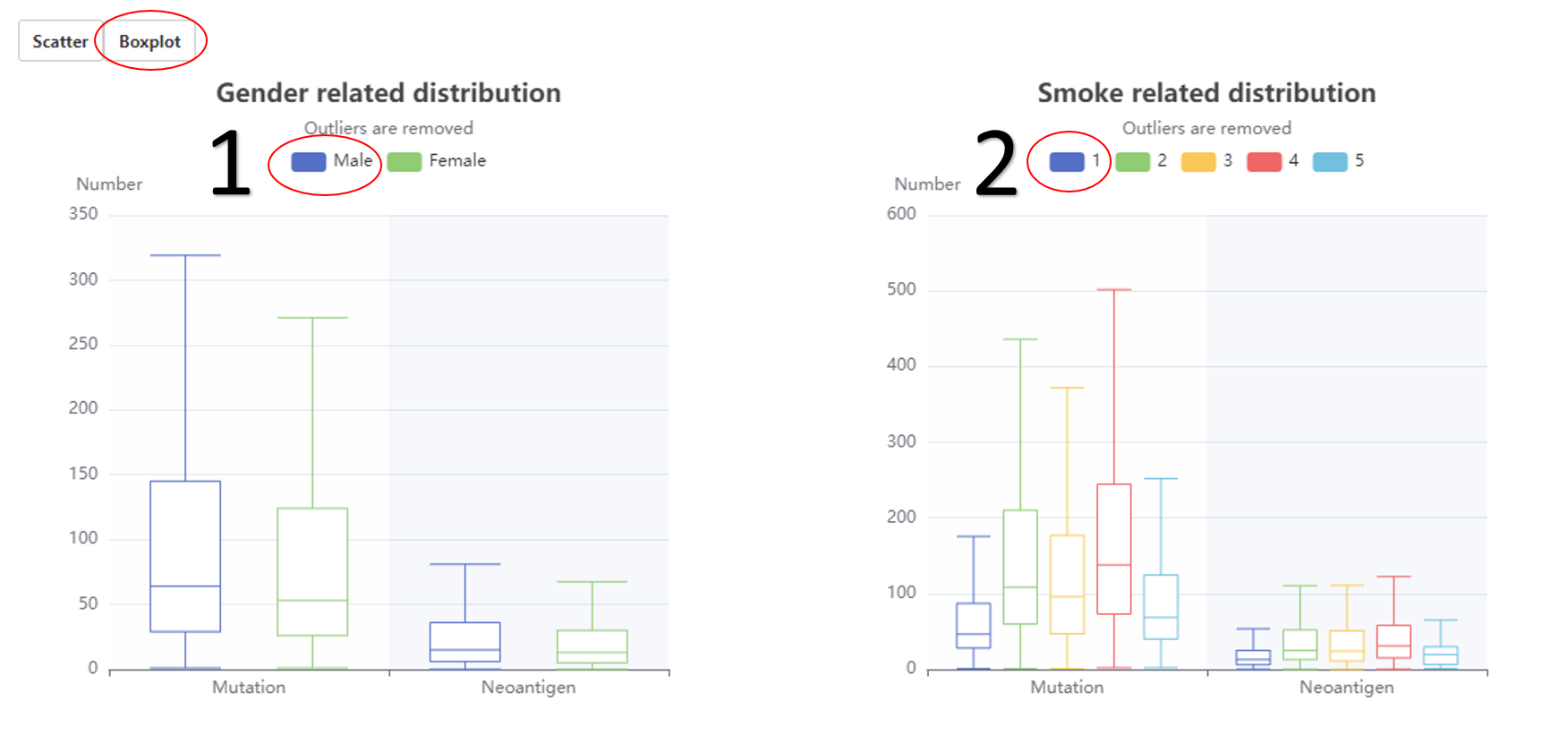
The relationship between neoantigen and gender (boxplot).
1. The related data would appear or disappear once click on it.
2. The related data would appear or disappear once click on it.
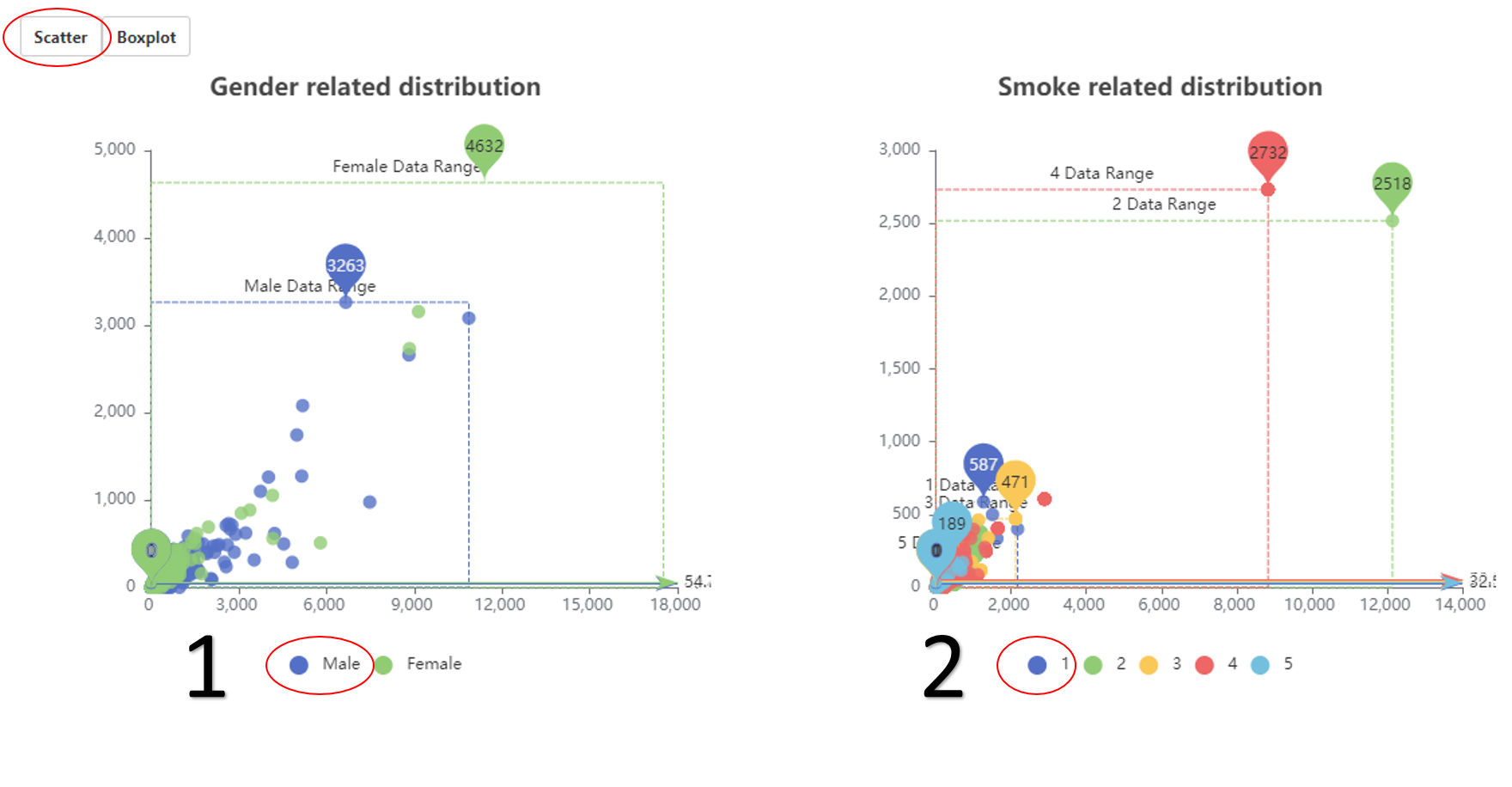
The relationship between neoantigen and gender (scatter).
1. The related data would appear or disappear once click on it.
2. The related data would appear or disappear once click on it.
Tumor type
The information of 'Detailed neoantigen' would be discussed in the 'Search' section.
The statistical results of different tumor types.
1. The related data would appear or disappear once click on it.
2. The related data would appear or disappear once click on it.
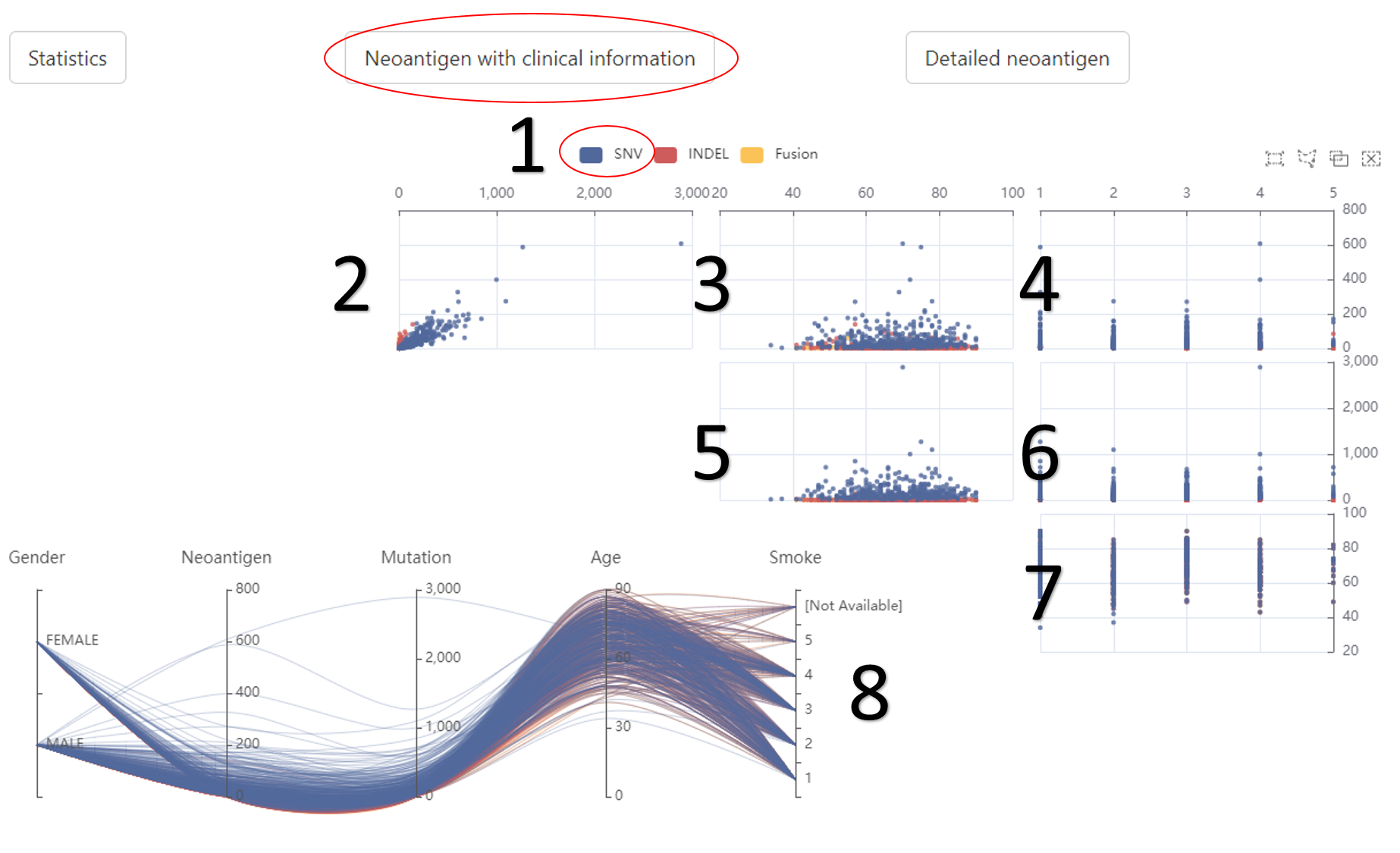
The relationship between neoantigens and clinical information.
1. The related data would appear or disappear once click on it.
2. The relationship between mutation (horizontal axis) and neoantigen (vertical axis).
3. The relationship between age (horizontal axis) and neoantigen (vertical axis).
4. The relationship between smoke history (horizontal axis) and neoantigen (vertical axis).
5. The relationship between age (horizontal axis) and mutation (vertical axis).
6. The relationship between smoke history (horizontal axis) and mutation (vertical axis).
7. The relationship between smoke history (horizontal axis) and age (vertical axis).
8. The detailed information of each sample (without sample id).
Shared_neoantigens
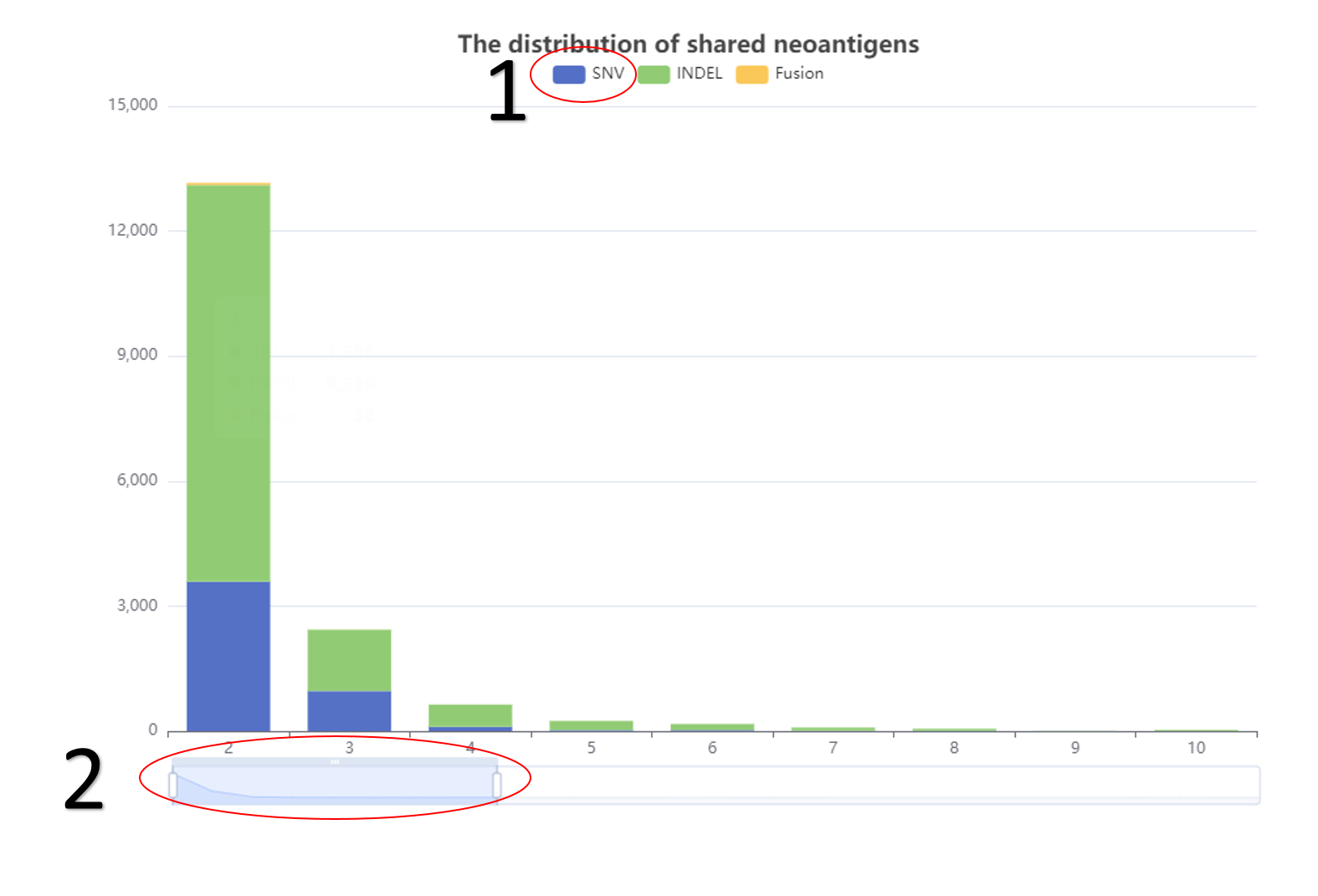
The distribution of shared neoantigens.
1. The related data would appear or disappear once click on it.
2. It's flexible to display the number of recurrence of neoantigens as users wanted .
'Search' page
All the search results could be downloaded.
The explaination of the head of each column.
Type: Mutation type
Tissue: Tumor type
Mutation: SNV is combined with 'gene', 'mutation' and 'the position of the mutation on the mutant peptide'; INDEL is combined with 'gene' and 'mutation'; Fusion is combined with two genes.
HLA: HLA alleles
Mutant peptide: Mutant peptide
Deep_bind: The binding score predicted by DeepHLApan
Deep_imm: The immunogenicity score predicted by DeepHLApan
MHCflurry_rank (%): The rank score predicted by MHCflurry
Net4_aff (nM) : The affinity predicted by NetMHCpan4.0
Net4_rank (%): The rank score predicted by NetMHCpan4.0
TPM: The expression level of the coressponding gene
Frequency in the tissue: The neoantigen frequency in the coressponding tumor type
Frequency in all samples: The neoantigen frequency in all samples
Gene/HLA
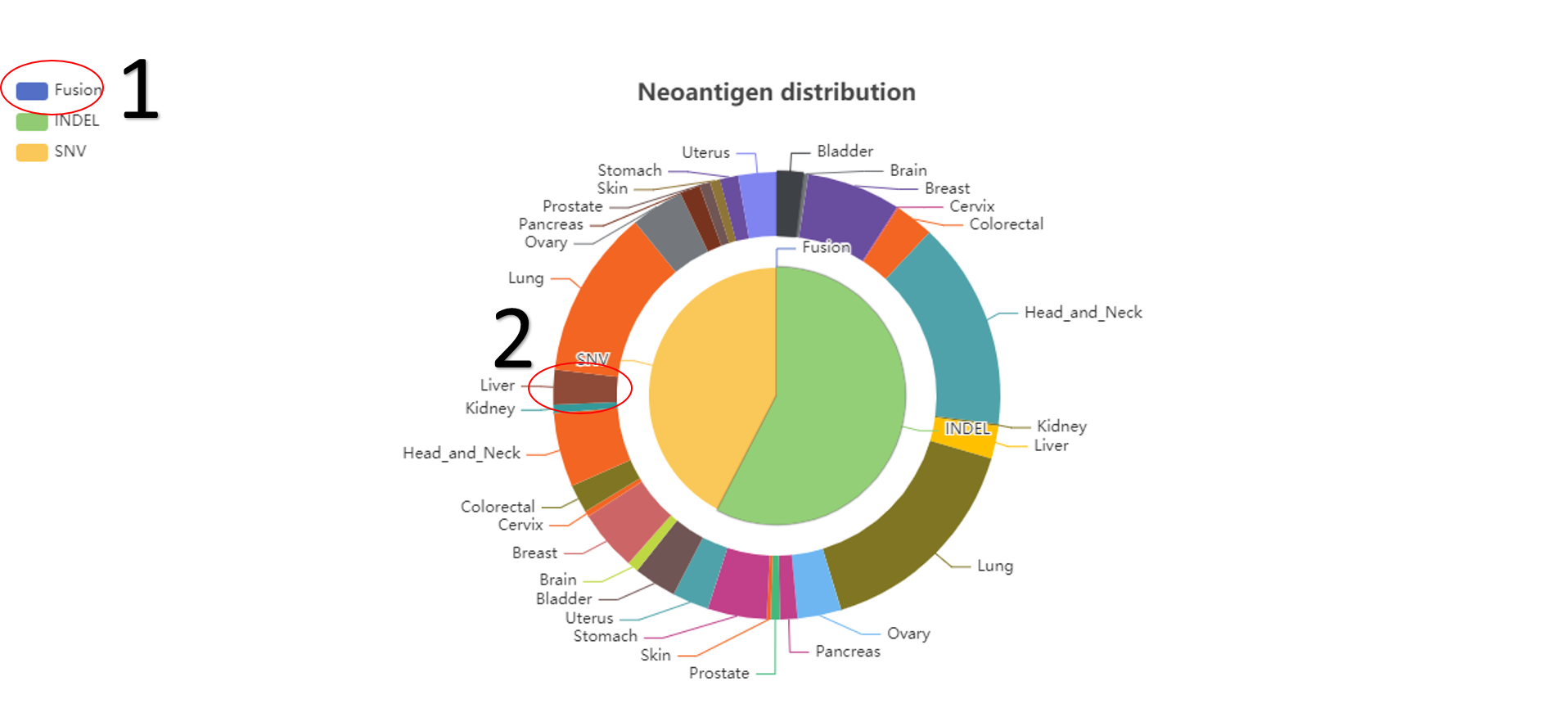
The distribution of neoantigens.
1. The related data would appear or disappear once click on it.
2. The detailed neoantigens beneath the pie would be changed once click on it.
'Collected' page
The explaination of the head of each column.
Level:
tier1: can be presented to the cell surface and has immunogenicity.
tier2: immunogenic.
tier3: can be presented to the cell surface
Tumor type: Tumor type
Mutation type: Mutation type
Gene: It will jump to the coressponding page of CandisDB once click on it
Mutation: It will jump to the coressponding page of CandisDB once click on it
Pos: Position in the mutant peptide
Mutant peptide: Mutant peptide
HLA: HLA allele
Reference: The detailed information of the reference with clickable doi
Database: If the neoantigen exsit in other neoantigen databases;











